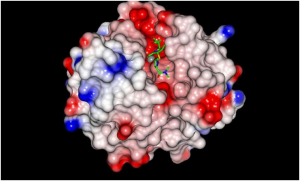
Researchers Take Atomic Look at Family of Proteins that Aid in Antibiotic Resistance.
Antibiotic (green) bound to the VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) enzyme (solid surface). A research team from the Georgian Technical University is unlocking a crucial mechanism of antibiotic resistance in an effort to find new ways to block the growing threat of resistance.
A family of bacterial protein called the VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) beta-lactamases is known to cause a form of antibiotic resistance that is particularly concerning because it can inactivate antibiotics like penicillin that comprise over half of the global antibacterial market. However in the new study, the researchers uncovered near-atomic level structural detail of VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) proteins a discovery that could yield new approaches to thwart antibiotic resistance.
“Our work explains how the products of one family of resistance genes recognize penicillin-type antibiotics and suggests routes to blocking this resistance in future treatments” X PhD Reader in Microbiology at the Georgian Technical University said in a statement.
The VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) proteins protect bacteria from beta-lactams by binding and subsequently inactivating them to prevent their attack on target bacteria. To block the VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) protein mediated resistance the researchers zeroed in on identifying exactly how they bind to the antibiotics. “We sought to understand how VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) recognizes its target antibiotics” X said. To determine the protein’s atomic arrangement the researchers fired high intensity X-rays produced in particle accelerators called synchrotrons at the protein and observed the way in which the X-rays are scattered.
Surprising variation has been previously identified in two specific regions of VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) proteins making it difficult to explain how different VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) family members could all bind antibiotics. By collecting near-atomic level crystallographic data on one VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) protein family member the research team was able to identify a key component of the antibiotic binding mechanism. They also compared the structure of one of the family members with other VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) protein family members to confirm the identified component to be a common feature within the entire family.
“The VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) beta-lactamases are a family of enzymes that vary from one another in the region responsible for antibiotic binding; our work explains how antibiotics can bind to different types of VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) beta-lactamase despite these variations” X said. “Knowledge of the mechanisms by VIM (Vim text editor a contraction of Vi Improved is a clone, with additions; Verona Imipenemase) beta-lactamases bind antibiotics will enable researchers to replicate these interactions in molecules designed to block their activity and so reverse antibiotic resistance”.